|
|
|
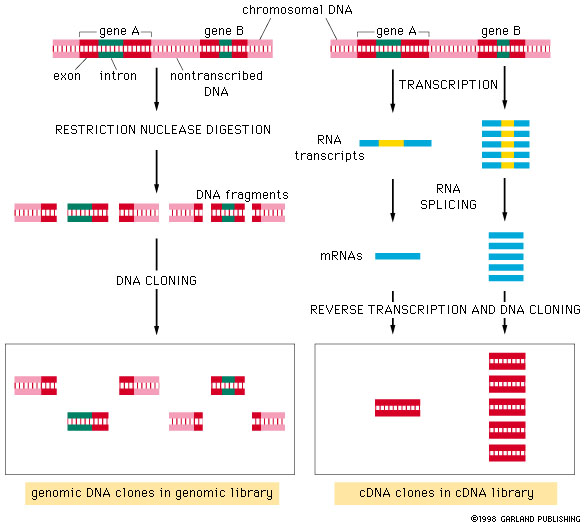
................. DNA libraries................
ln(1-P)
N = -----------
ln(1 -1/n)
where N is the number of clones needed to have a probability, P, of finding any particular sequence in the library, when the ratio of the genome size to the average cloned fragment size is n. The same formula applies to cDNA libraries, but 1/n is defined as the fractional proportion that a specific mRNA is of the total mRNA.

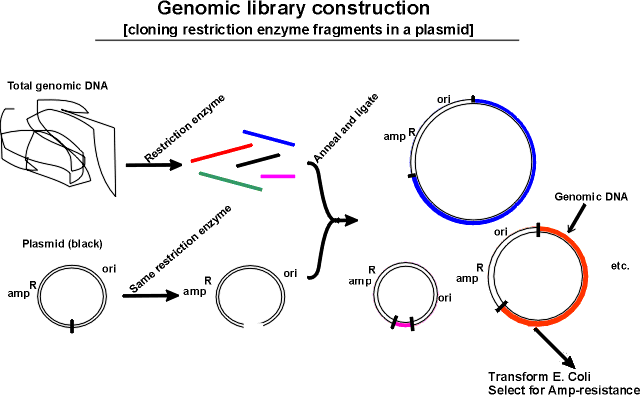
For prokaryotes (smaller genomes) - viable to make gene libraries in plasmids. Plasmid inserts normally ~5-10 kb, so only need a few thousand recombinants for a representative library. For eukaryotes (larger genomes), really need vectors that can hold much larger insert DNA fragments.
Alternative vector systems:
| Approximate maximum length of DNA that can be cloned into vectors | |
| Vector type | |
| Plasmid | |
| lambda phage | |
| Cosmid | |
| P1 phage | |
| BAC (bacterial artificial chromosome) | |
| YAC (yeast artificial chromosome |
Bacteriophage lambda - much more efficient infection of E. coli than achieved
by plasmid transformation
P1- E. coli virus capable of packaging
large DNA segments.
Bacteriophage lambda
Genes involved in lysogeny and certain other genes irrelevant for lytic growth can be deleted from the viral genome, and replaced by other DNA sequences of interest. This is the basis of the use of phage lambda as a vector. Insert size is limited to ~ 25 kb due to the requirement that the DNA has to fit into the phage head.
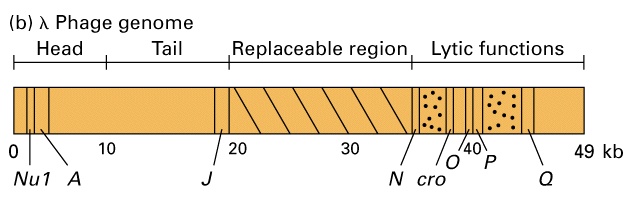
Fig 7-10b Lodish et al, 4th ed
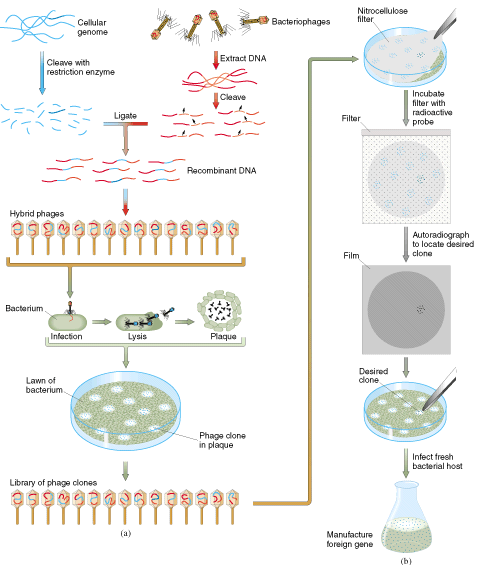
Making a genomic library in
bacteriophage lambda vector:
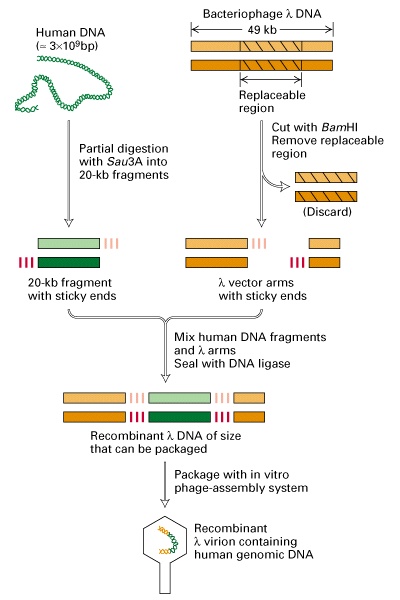

Phage lambda bind to receptors on the surface of E. coli and injects DNA. Infection by lambda is much more efficient than plasmid transformation - can get ~ 109 plaques per microgram of DNA, vs ~ 106 colonies per microgram of plasmid DNA
As the bacterial cells lyse, a plaque forms, which spreads as more cells become infected and lyse. The plaque contains the infectious viral particles or virions and each one represents an individual lambda clone (similar to a bacterial colony representing an individual plasmid clone). The appearance is of a clear circle in a 'cloud' of bacterial cells (the bacterial 'lawn'). If incubated for long periods, these circles will continue to grow (containing more and more viral particles) and spread until all bacterial cells are wiped out.
To pick a single lambda clone, the plaque is 'punched' out from the agar plate and stored in a holding solution. This can then be used to infect more bacterial cells and to replicate more of the recombinant lambda DNA.
Vectors for making libraries with larger inserts
Cosmid libraries are used for cloning genes with large introns and for sequencing larger chunks of the genome. Cosmid vectors are hybrids of plasmid and bacteriophage lambda DNA (a small ~5 kb plasmid containing the plasmid origin of replication (ori), an antibiotic resistance gene such as ampr and a suitable restriction site for cloning along with the COS sequence from phage l DNA). Because of the COS sequence, cosmid recombinants can be packaged into viral particles (allowing high efficiency transformation). Since most of the l DNA has been discarded, it can be replaced by the DNA of interest, so long as it doesn't exceed the 50 kb limit for packaging into the viral head. The insert sizes are of the order of 35-45 kb. Since the recombinant DNA does not encode any lambda proteins, cosmids do not form viral particles (or plaques) but rather forms large circular plasmids and the colonies that arise can be selected on antibiotic plates, like other plasmid DNA transformants. Cosmid clones can be manipulated similarly, allowing ease of plasmid isolation (see previous lecture). Since many eukaryotic genes are on the order of 30 - 40 kb, the likelihood of obtaining a DNA clone containing the entire gene sequence is increased significantly when using a cosmid library.
YAC libraries are used for cloning very large DNA fragments (of more than 1 Mb), and are useful for cloning large genes (such as the 250 kb cystic fibrosis gene) and for creating libraries of large overlapping clones, such as for individual chromosomes isolated from organisms (chromosomal libraries). These have been used extensively for mapping genomes of complex organisms (e.g. Homo sapiens).For example, Trp1 mutants can't make tryptophan so can only grow on media supplemented with tryptophan. If the mutant strain is transformed with a YAC containing an intact Trp1 gene, then this will compensate for the inactive gene (complement) and the transfected cell is able to grow on media lacking tryptophan.
YACs
are not used as extensively anymore because of inherent problems. For
instance, YAC clones can contain
non-contiguous segments of the genome. This means that 2 or more DNA fragments
from separate parts of the genome can be integrated into an individual YAC
(because they are able to support rather large inserts). A second problem is
that YACs are unstable and frequently lose
parts of the DNA during propagation.
BAC libraries are also used for cloning
very large DNA fragments and have been particularly useful for sequencing
large genomes. BACs are bacterial
artificial chromosomes, and are based on a naturally occurring
large bacterial plasmid, the F-factor. BAC vectors
can accommodate DNA inserts up to 300 kb, still fairly respectable when
needing to clone large genes or map and sequence complex genomes.
BACs have several advantages over YACs, which means they are used more extensively now. Most of the human genome has been sequenced using BAC rather than YAC clones
images sources: they are selected from many sites.